How long-read sequencing will transform neuroscience
New technology that delivers much more than a simple DNA sequence could have a major impact on brain research, enabling researchers to study transcript diversity, imprinting and more.

Genomics is in the midst of a major transformation, thanks to the advent of technology that can read sequences up to tens of thousands of base pairs long. Known as long-read sequencing, this method brings with it a host of new capabilities: Unlike the current standard, short-read sequencing, it can easily generate whole-genome sequences from scratch, without the need for a reference genome. It can sequence RNA directly and detect chemical modifications involved in gene expression. These applications are particularly exciting for the field of neuroscience. Long-read sequencing could offer a much richer picture of the diversity of RNA transcripts in the brain, for example, revealing how this heterogeneity varies with brain region and in different diseases.
Short-read sequencing revolutionized genomics in many ways — it played a critical role in the Human Genome Project, and it brought on the “-Seq” epoch, forming the basis for whole-exome sequencing, whole-genome sequencing and RNA sequencing, as well as methods for identifying transcription factor binding sites, known as ChIP-sequencing, and for detecting chromosome accessibility, known as ATAC-sequencing.
Still, short-read sequencing has its limits. Because it typically reads sequences of only 150 base pairs or shorter, these pieces must be computationally stitched together to create a larger stretch. It can’t sequence RNA directly, and it has trouble with repetitive sequences of DNA, which can play an important role in the brain — highly repetitive protocadherins, for example, come together in different combinations to control neural signaling.
These limitations led researchers to develop strategies for sequencing full-length transcripts. Though early methods struggled with high error rates, recent breakthroughs have dramatically improved accuracy. As of November 2023, the two most commonly used long-read sequencing technologies are Pacific Biosciences HiFi, which can generate sequence reads up to 20,000 base pairs long with greater than 99.999 percent accuracy, and Oxford Nanopore technology, which can generate megabase-length sequence reads with more than 99.9 percent accuracy. The growing popularity of long reads will likely draw other competitors to this space.
Long-read sequencing is still significantly more expensive than short-read technology. The cost is falling rapidly — 10-fold in the past year — but needs to drop 2- to 3-fold more to compete with short reads. Long reads give much more information, however, so the technology is already cost effective in many ways.
B
ecause long-read technology can sequence RNA directly, it can reveal alternative editing to RNA transcripts, something that has been difficult to do accurately with short-read methods. Elegant new work in this area is shining a light on the diversity of RNA transcripts, known as isoforms, in different brain regions and cell types. In particular, researchers will get a more complete picture of genes that may have many isoforms, such as CACNA1C, a calcium channel gene linked to a variety of neurological and developmental conditions. Future applications include subcellular transcriptome sequencing to examine RNA in different compartments of neurons, such as at the synapse and in glia.Unlike short-read methods, long-read sequencing can directly detect base modifications that regulate gene expression, such as 5-Methylcytosine DNA methylation and N6-Methyladenosine in mRNA. It can also directly determine on which chromosome — maternal or paternal — a variant resides, which can be important for a variety of developmental conditions. Angelman syndrome, for example, is caused by changes in the maternal but not paternal copy of the UBE3A gene. Long-read sequencing might even be used to explore interactions related to DNA’s 3D structure, which may play a role in gene expression. A technology called Pore-C, for example, can examine higher-order genome contacts — a capability that will likely be useful for understanding regions of the genome containing gene families important for the nervous system, such as the alpha, beta and gamma protocadherins.
Long-read sequencing is also helping researchers better characterize in-vitro and in-vivo systems commonly used in neuroscience, including induced-pluripotent-stem-cell-derived neurons; immortalized cell lines; neuronal and glial cultures; and C. elegans, Drosophila, mouse, rat and zebrafish models. Reference genomes, epigenomes and transcriptomes exist for some of these systems. But the field would benefit from deeper characterization of mutant systems with disrupted nervous systems and those engineered to mirror human neurological disease. Ongoing projects include the International Mouse Phenotyping Consortium, the Drosophila Genetic Reference Panel, the Caenorhabditis Genetics Center, the Simons Foundation Rat Models and the Zebrafish International Resource Center. Several groups are performing long-read sequencing on these lines to provide deeper genomic, epigenomic and transcriptomic resources and insights to the broader research community. Projects aiming to perform long-read sequencing on all organisms on Earth may even lead to new model organisms.
All of these capabilities will also help advance the discovery of the genetic underpinnings of neurodevelopmental and neurodegenerative conditions. Clinical genetic testing is currently limited by inferior short-read genomic technology. Use of long-read sequencing in the research space is demonstrating what we have been missing with short reads, including direct sequencing of expanded DNA repeats, identification of which chromosome a variant resides on (known in technical terms as physical phase status of variation) and methylation status, all of which can be important when studying the genetics of disease. Because this technology enables custom de novo assembly of a person’s genome to piece together complex variation, obviating the need for a reference genome, it also avoids the biases reference-based analyses can incur. Long reads have been applied in the discovery of relevant variation in Alzheimer’s disease, autism, epilepsy, motor neuron syndrome, Parkinson’s disease, spastic paraplegias and spinocerebellar ataxia, to name a few.
Going forward, the field needs to streamline and optimize bioinformatic and computational workflows used to analyze the data and generate a large resource of long-read sequencing from healthy people. Each is the focus of intense research. The All of Us research program and the Human Pangenome Reference Consortium are both building long-read genomes from people without disease, for example. And long-read sequencing also needs to get faster, although it is making major strides in speed — last year, Stanford University researchers set the Guinness World Record for ultra-rapid long-read sequencing, reading a person’s genome in just five hours and two minutes.

Washington University School of Medicine
Explore more from The Transmitter
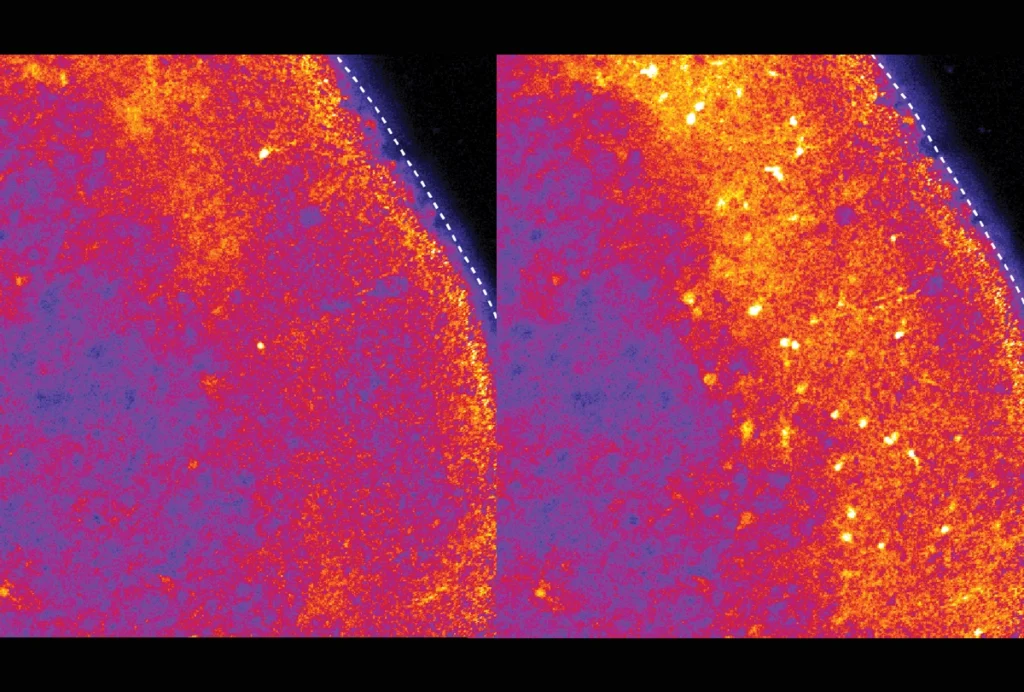
RNA drug corrects calcium signaling in chimeric model of Timothy syndrome