THIS ARTICLE IS MORE THAN FIVE YEARS OLD
This article is more than five years old. Autism research — and science in general — is constantly evolving, so older articles may contain information or theories that have been reevaluated since their original publication date.


The past decade has seen an exponential increase in our understanding of the genetic underpinnings of autism and other neurodevelopmental conditions. Despite this progress, we understand little about the cellular and circuit-level dysfunction in the brain that is associated with these conditions. Part of this difficulty arises from the complexity of the human nervous system at every level, from molecules to systems.
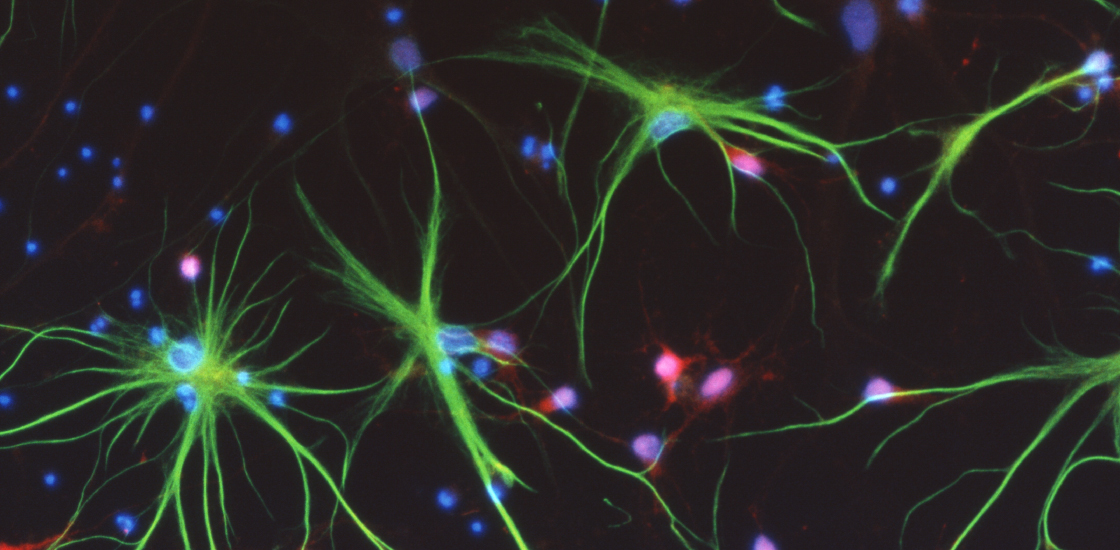
Among the most ubiquitous and elusive human brain cells are astrocytes — star-shaped cells that support neurons in various ways. Human astrocytes and other kinds of glial cells — a broader class of brain cells that protect, anchor and insulate neurons — offer a new constellation of drug targets that simply don’t exist in current animal models. We need new techniques to study human astrocytes and their potential role in autism and other conditions.
For example, we still don’t know how astrocytes and other brain cells function and interact with each other during typical and altered development in people. Emerging technologies — ranging from deep sequencing to culturing populations of neural cells as three-dimensional ‘mini-brains’ — are beginning to produce evidence of human cellular and molecular uniqueness. But we have a long way to go to uncover the many mysteries of astrocytes in people.
Astrocytes are implicated in modulating most — if not all — aspects of neuronal development and function. They take in and secrete molecules important for both developing and maintaining synapses (connection hubs between neurons), which often appear to be altered in experimental models of autism.
Large genetic studies over the past few years have uncovered numerous genes that are implicated in autism, and many of these are expressed in astrocytes. Increasing evidence from genetic mouse models also suggests that astrocyte development is disrupted in numerous neurodevelopmental conditions, including autism1.
Using the emerging technologies, we can begin to address some of the major outstanding questions in autism, such as: How do astrocyte cellular changes lead to cognitive impairment? Are there ‘good’ and ‘bad’ astrocytes in conditions such as autism? If so, how can we take advantage of this information? How can we target astrocytes for therapy?
Amplified role:
As early as the 1890s, scientists observed that astrocytes come in a range of types, but that those in people differ in shape and function from those in mice and nonhuman primates. It’s only over the past decade that some teams have detailed these differences, however2,3,4. For example, single human astrocytes reach dramatically longer distances in the brain and propagate calcium signals faster than astrocytes in other species. These features suggest an amplified role for human astrocytes in brain function.
Using large datasets, researchers have also found that, unlike the gene expression signatures of neurons, astrocyte gene expression in humans is quite different from that in mice. This finding provides yet more evidence that astrocyte evolution selectively diverged among mammalian species5.
What was the selective pressure in hominids that led to these large, more complex and molecularly distinct astrocytes? And do these unique features of astrocytes make people particularly susceptible to autism-related traits when development is perturbed?
It is unclear whether at a particular point in development certain intrinsic factors, such as proteins that control gene expression, kick in to endow astrocytes with their distinct functions6. Studies in the past two years suggest that astrocytes need molecular signals from nearby neurons in order to take on a particular identity as a distinct subtype of astrocytes in the cerebellum7. Other studies demonstrate that proteins that control gene expression, and thereby regulate cell identity during development, play important roles in specifying astrocyte subtypes8.
This diversity means that we may need to tailor treatments to the particular type of astrocytes affected. For example, astrocytes in one part of the cerebral cortex — the outer layer of the brain — may respond differently to drug treatment than astrocytes in another part.
The studies explaining the origins of the many types of astrocytes were all done in animal models, however. So far, drugs that have successfully targeted brain function in mice have generally failed to show efficacy in clinical trials. Given the many differences between human astrocytes and mouse astrocytes, we need to create better model systems involving human glia to speed new drug discovery.
Cellular solutions:
One way to chart the unique features of human astrocytes is to culture them from brain tissue taken during surgeries. When directly compared with rat and mouse astrocytes cultured in the same way, the differences in size and gene expression become apparent9.
Producing a ‘humanized’ model of somewhat intact neural circuits can also facilitate studies of astrocytes, as these models provide a better environment to observe cell function than traditional mouse models. Already, researchers have grafted precursors of human astrocytes into the nervous system of a mouse to study cognitive function10. They have also successfully replaced astrocytes in the mouse spinal cord with human cells to test treatments for spinal cord injury and to investigate the role of human astrocytes in neurodegenerative disease11,12.
Researchers can also use three-dimensional mini-brains to study human-specific features of astrocytes. These cultures recapitulate the inside-out development of the human cerebral cortex and generate astrocytes around the same time the brain does, potentially allowing us to uncover both early and late roles for human astrocytes in brain development13.
To understand how astrocytes are altered in conditions such as autism, one could take cells from an individual with the condition and generate glia from stem cells. We could also introduce autism-linked mutations into these cells using recent genetic engineering technology. Already, we are starting to see the early potential of these methods.
In a paper published last year, we derived astrocytes from stem cells created from people with Costello syndrome, an autism-related condition. We found that these astrocytes mature faster and also enhance early maturation of neighboring neurons14. Our findings suggest that targeting human astrocytes in addition to neurons may be a viable route for developing treatments for autism and related conditions.
Eric M. Ullian is associate professor of neuroscience at the University of California, San Francisco School of Medicine, where he leads a lab in the ophthalmology department. Robert Krencik is a postdoctoral researcher in his lab.
By joining the discussion, you agree to our privacy policy.