THIS ARTICLE IS MORE THAN FIVE YEARS OLD
This article is more than five years old. Autism research — and science in general — is constantly evolving, so older articles may contain information or theories that have been reevaluated since their original publication date.
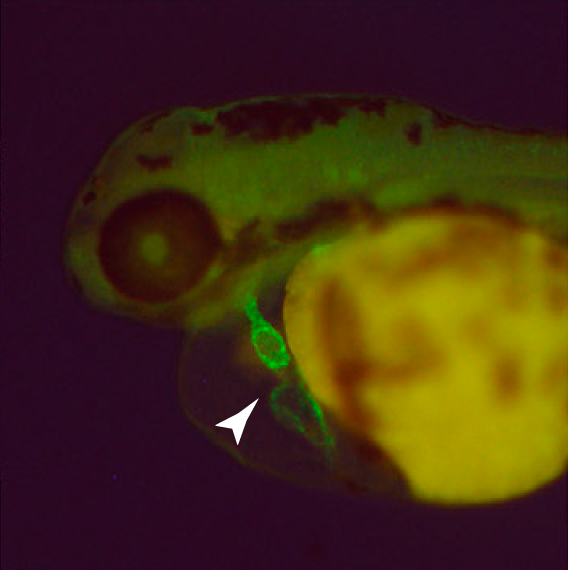
Green gene: Researchers can easily see an edited gene in the zebrafish heart (arrow) because they tagged it with a green fluorescent protein.
A new gene-editing system allows researchers to rewrite lengthy stretches of DNA in zebrafish and identify the mutant fish with ease. Researchers can later remove the edited sequences from the fish1.
The new technique harnesses the gene-editing power of either of two types of ‘protein scissors:’ CRISPR-CAS9 or TALENs. Researchers can use these tools to cut out a tiny stretch of zebrafish DNA and swap in an edited version of the same sequence.
The system, described 21 March in Developmental Cell, delivers the edited material on a circular piece of DNA called a plasmid, which can hold long sequences encoding entire genes as well as elements such as fluorescent tags.
The researchers made plasmids containing the edited versions of genes for skin pigmentation or heart function, and outfitted each plasmid with the blueprint for a fluorescent protein tag. Because zebrafish embryos are transparent, those that integrate a plasmid into their genomes glow under ultraviolet light.
This color cue allows researchers to easily detect the roughly 5 to 15 percent of embryos that express the edited gene. Previously, scientists had to sequence thousands of fish to find the few that had incorporated a new gene.
The plasmid also encodes a switch that ejects all or part of the swapped-in material. Inserting DNA ‘landing strips’ for a cutting enzyme called CRE allows scientists to remove all or part of a swapped-in sequence after injecting the enzyme.
Color clues:
Researchers tested the new system in fertilized eggs carrying a mutation in GOL, a gene that gives zebrafish their dark pigment. Using the TALENs tool, they snipped out the GOL mutation and used a plasmid to swap in a functional version of the gene with a fluorescent tag. This turned the mutant embryos from light gold to dark brown and caused them to glow green under ultraviolet light.
The fluorescent tag could be used in this way to speed studies of mutations that do not have obvious effects, the scientists say.
They also tested the system’s ability to turn genes off during certain stages of development. Using recently fertilized zebrafish eggs, they used TALENs to target a gene called KCNH6A, which plays a crucial role in the heart.
They were able to snip out KCNH6A using CRE and replace it with a nearly identical sequence containing a fluorescent tag. They then cut out the replacement gene so that the fish carrying it no longer glowed; these fish also developed lethal heart defects.
The system cuts down the time and resources needed to identify fish carrying edited genes. It could also help to reveal the effects of autism-linked mutations during specific phases of zebrafish development.
By joining the discussion, you agree to our privacy policy.