A new tool uses a customizable RNA molecule to identify and manipulate individual cell types in living animals and human brain tissue. It could help researchers test how different cell types contribute to autism.
To label or control select cells, researchers typically use a virus engineered to carry a gene of interest along with a specialized enhancer — a stretch of nucleotides that can activate a nearby gene, but only when it binds itself to proteins present in the target cells. After the virus is injected into an animal or a tissue sample, only those target cells express the desired gene. But the method is limited because few cell-specific enhancers have been identified.
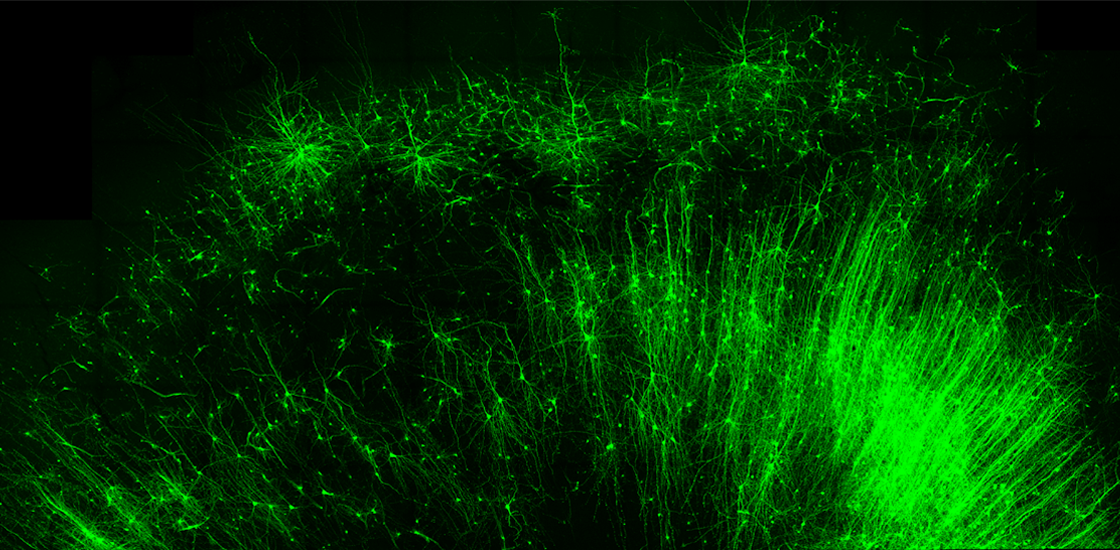
Target cells also display distinct patterns of gene expression — an RNA fingerprint. The new approach, called CellREADR, harnesses cell-specific RNAs to sidestep the need for an enhancer and deliver an RNA copy of a gene directly into target cells.
“Our new technology is a fundamental departure from previous DNA-based approaches,” says lead investigator Josh Huang, professor of neurobiology at Duke University in Durham, North Carolina.
Huang’s team engineered a probe made up of three key RNA components: a ‘sensor’ sequence that binds to cell-specific messenger RNA (mRNA), the coding sequence for the gene of interest and an ‘inhibitor’ sequence that prevents that gene’s expression. When the sensor binds mRNA, the resulting double-stranded structure activates a naturally occurring editing enzyme called ADAR, which replaces letters in the probe.
Specifically, ADAR edits and inactivates the inhibitory sequence, triggering the synthesis of the protein encoded by the probe. The inhibitory sequence remains intact in cells that don’t express the mRNA, so no protein is built there. And because ADAR is present in all tissues across all animal species, it can be adapted to different disease models, the researchers say.
It could be a “game changer” for autism research, says Karun Singh, senior scientist at the Krembil Research Institute at the University Health Network in Toronto, Canada, who was not involved in the study. Researchers could target CellREADR to autism-linked genes to monitor how strongly they are expressed in different cell types, he says.
What’s more, advances in single-cell brain atlases will likely uncover a diversity of brain cell types, some of which may be important in autism, Singh says. Detection using RNA-based approaches means researchers aren’t hampered by the availability of good antibodies, he adds.
T
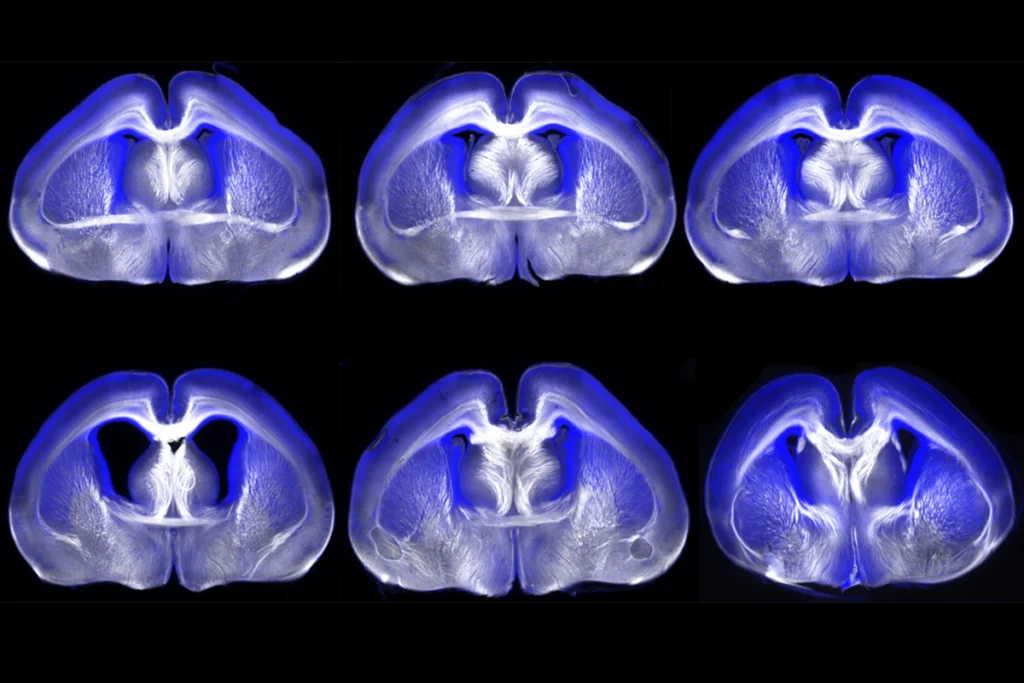
They then modified the probe to instead code for a light-activated version of an ion channel protein and injected it into a brain region involved in movement. Light stimulation of the brain activated CTIP2-specific glutamatergic neurons in the region and caused the mice to move their front paws, highlighting the tool’s potential for controlling cell function in animals.
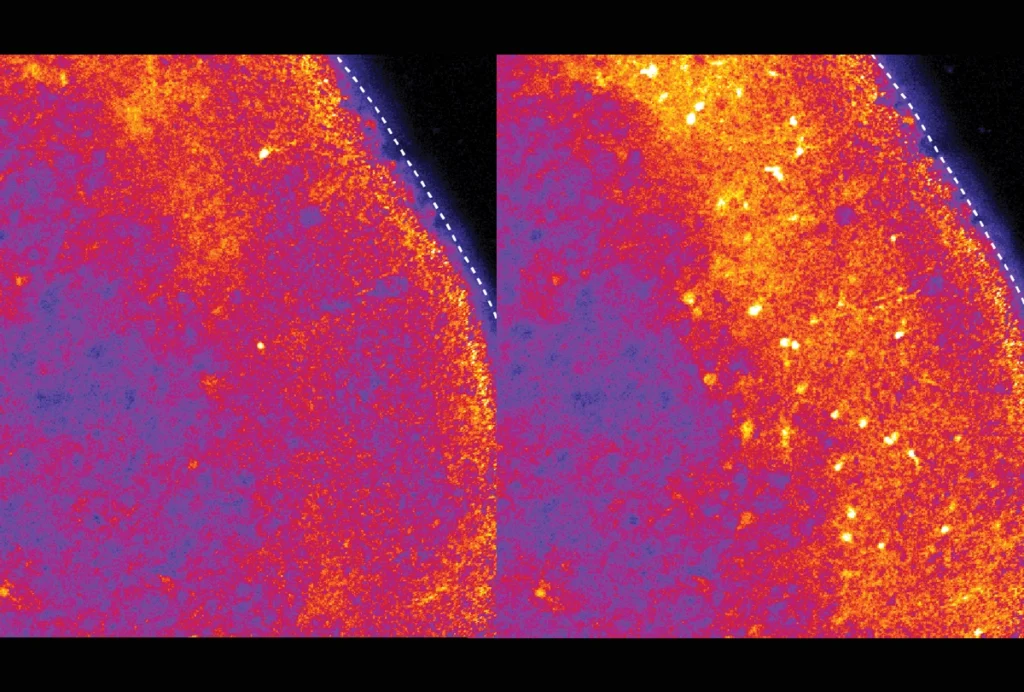
The technique also worked in cortical brain tissue removed from people with epilepsy during surgery. A probe designed to express a fluorescent protein when it senses mRNA for FOXP2, an autism-linked gene involved in learning language, lit up neurons responsible for learning and memory, the researchers found. The findings appeared in October in Nature.
The new tool could also be adapted for therapeutic purposes, Huang says. Some forms of autism stem from mutations acting very early in development, making it “almost impossible to have an impact by correcting the genes.” But different mutations converge on the same brain circuits, which scientists can target by manipulating the cell types that make up that pathway, he says.