THIS ARTICLE IS MORE THAN FIVE YEARS OLD
This article is more than five years old. Autism research — and science in general — is constantly evolving, so older articles may contain information or theories that have been reevaluated since their original publication date.
A new technique classifies neurons by surveying chemical tags that turn genes on or off on the neurons’ DNA1. The approach represents a new way to chart the brain’s cellular diversity. It could reveal how patterns of chemical tags known as methyl groups are altered in autism.
Methyl groups bind to the DNA base cytosine. Patterns of methylation can be inherited, but they can also change in response to environmental factors, such as exposure in the womb to stress hormones or to the mother’s diet. Studies have reported altered methylation patterns in postmortem brains of people with autism.
Methylation patterns also vary by cell type. In a new study, published 11 August in Science, researchers classified neurons from mouse and human brain tissue by their methylation patterns.
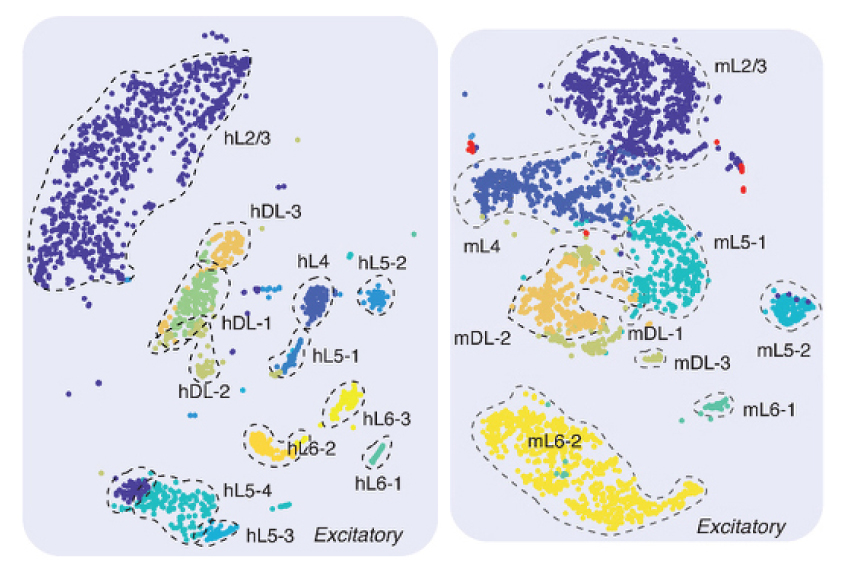
The researchers looked at cells from specific layers of the brain’s outer shell, the cerebral cortex. They used a chemical cocktail to isolate the cells’ nuclei, and placed a single nucleus in each well of a 384-well plate. They then treated the nuclei with a chemical that converts cytosines without methyl groups to the RNA base uracil. They sequenced the DNA to pinpoint the remaining cytosines, yielding a map of every methyl group.
Playing tag:
The researchers applied the approach to 3,377 mouse neurons and 2,784 human neurons, revealing 16 types of mouse neurons and 21 types of human neurons, each with a distinct pattern of methyl markings.
The sequencing step allowed the researchers to determine which genes are under the control of methyl groups in each type of neuron. They were then able to deduce whether the genes were turned on or off based on the placement of the tags within a gene or in the surrounding regulatory regions.
Many of the methylation patterns correspond with known patterns of gene expression in specific types of neurons. But two methyl group arrangements point to unique gene-expression profiles, hinting at two previously unrecognized types of neurons.
One of the new types boosts brain signaling in mice, and the other dampens brain signaling in people.
By joining the discussion, you agree to our privacy policy.