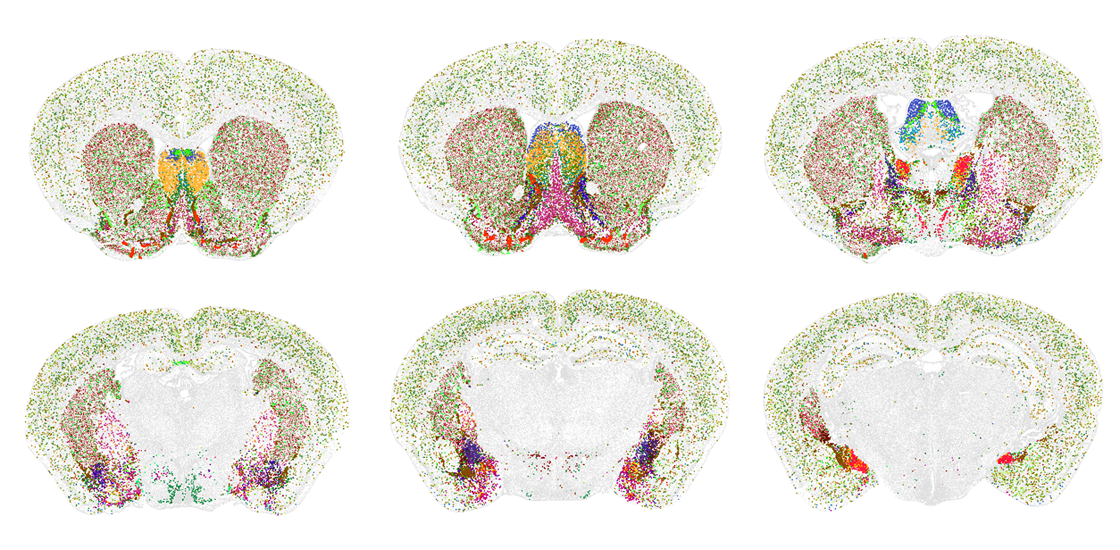
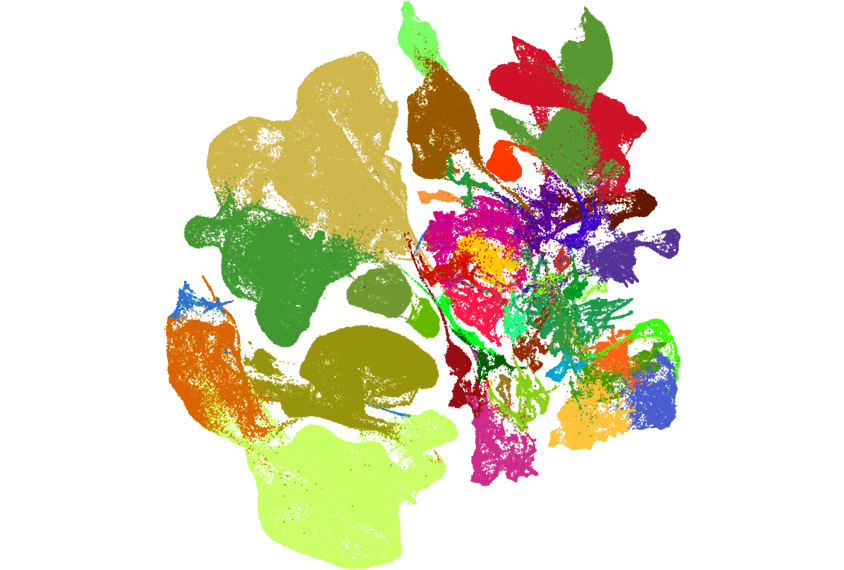
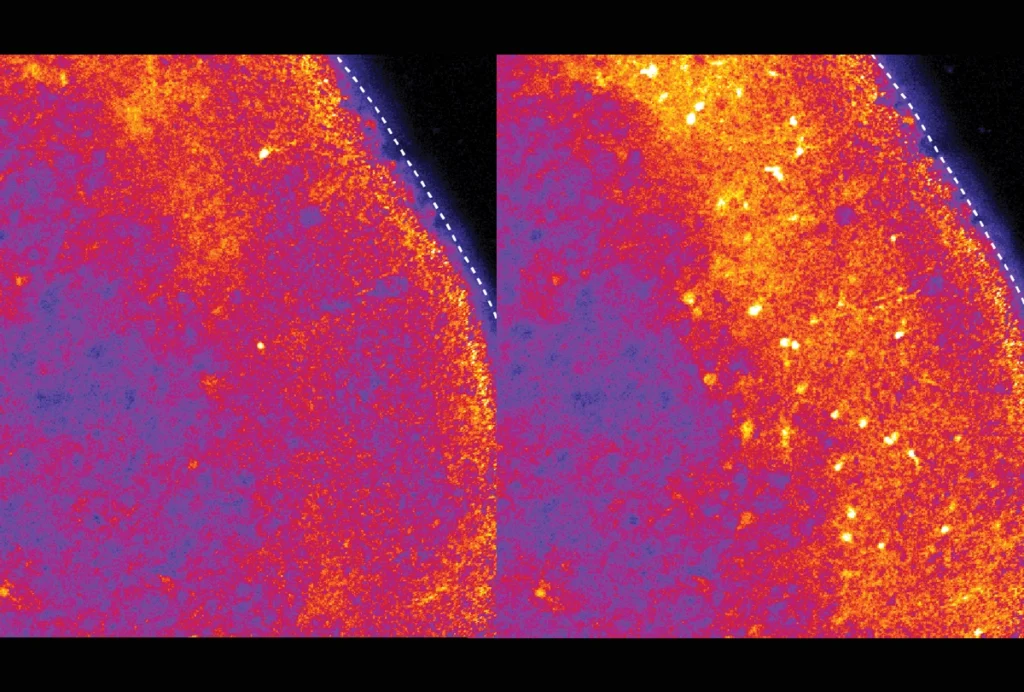
Two new atlases catalog the location and type of all cells across the adult mouse brain — including many that have never before been identified, according to two new unpublished studies.
The resources represent “a culmination of landmark efforts” to try to establish a census of cell types, says Tomasz Nowakowksi, associate professor of neurological surgery at the University of California, San Francisco, who was not involved in the work. “It’s presumably something that has been a dream since the days of Santiago Ramon y Cajal.”
In an effort spanning more than 10 years, and using different approaches, two teams of researchers each identified about 5,000 distinct clusters that represent different kinds of cells.
The “tremendous diversity” they observed was surprising, says Hongkui Zeng, executive vice president and director of the Allen Institute for Brain Science in Seattle, Washington, who led the work on one of the new atlases. Some of the clusters map to known cell types, Zeng says, but others “we never heard about before.”
That new information will help guide future studies, Nowakowski says. “Imagine that you’re trying to sail across the ocean, and you don’t really have a map,” he says. “That’s exactly what’s been happening with neuroscience for the past 100 years.”
T
The team also located the RNA molecules within the same slices of brain tissue using a method called multiplexed error-robust fluorescence in situ hybridization (MERFISH), which associates each RNA with a barcode linked to a particular spot. They then flagged which cell-type clusters appeared at a given spatial location using the previously obtained molecular handles.