THIS ARTICLE IS MORE THAN FIVE YEARS OLD
This article is more than five years old. Autism research — and science in general — is constantly evolving, so older articles may contain information or theories that have been reevaluated since their original publication date.
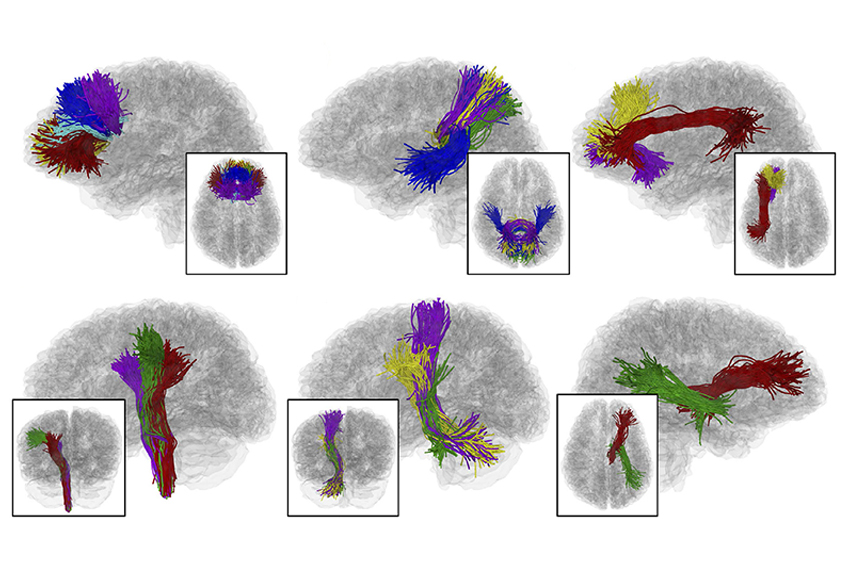
A new software package identifies nerve-fiber bundles in imaging data and maps the microstructure, length and trajectory of those ‘tracts’1. The software can spot differences between people with autism and controls.
Scientists can visualize neural connections using diffusion magnetic resonance imaging (MRI). This technique tracks the direction and speed of water molecules as they flow through the brain, and then computer software translates those measurements into pictures of the brain’s wiring.
Scientists can use diffusion MRI to test, for example, whether specific fiber tracts are unusual in autism. But this hypothesis-driven approach can miss alterations in other tracts. The new method automates the analysis to objectively illuminate differences in multiple fiber tracts.
It uses brain-imaging data from people with and without a condition such as autism to build a basic template outlining white-matter tracts in the brain. The software randomly samples 5,000 nerve fibers — about 1.5 percent of the total in a brain — from each of the scans. It excludes fibers that deviate substantially from the norm.
Then a machine-learning algorithm compares the fibers in each brain scan with the template and identifies the differences in each. The comparison reveals tracts with features that distinguish people with a condition. It can also classify whether individual scans came from a person with the condition.
Bridging differences:
Researchers tested the method on scans from 149 children — 70 with autism and 79 controls — from the Center for Autism Research at the Children’s Hospital of Philadelphia. The method accurately classified whether a brain scan came from a child with autism or a control 78 percent of the time.
It also showed that white-matter tracts in children with autism tend to be structurally weaker than those in controls. The differences between people with autism and controls are particularly prominent in several regions, including the corpus callosum, which bridges the brain’s hemispheres, and the cerebellum, which regulates movement.
White-matter tracts that run through the ventral diencephalon, a deep brain region that contains the hypothalamus — a structure that regulates body temperature and the day-night clock — are larger in children with autism than in controls. And researchers could use the method to identify other aberrations in nerve tracts specific to autism.
Scientists can download the software to analyze their own diffusion MRI datasets. The method was described 25 October in NeuroImage.
By joining the discussion, you agree to our privacy policy.