A mega-map of protein interactions provides a picture of how autism-linked mutations might disrupt business as usual in the cell, a new preprint reports.
More than 200 genes are strongly associated with autism, but how mutations in those genes lead to the many different outcomes that are diagnosed as autism is largely unknown. The new work builds on studies that have begun to systematically probe protein-level interactions to chart the effects of these mutations in the cell.
The research features the largest set of protein-protein interaction maps for autism-linked genes to date. The interactions identified, most of which had never been investigated before, group into multiple clusters based on overlapping function.
“We know that genes don’t work alone — they work in protein networks, and they form protein complexes,” says study investigator Belinda Wang, a postdoctoral researcher in Jeremy Willsey’s lab at the University of California, San Francisco. She and her colleagues used mass spectrometry to create large-scale models to investigate how specific mutations in the genes affect protein networks.
Identifying the overlapping molecular functions within these maps could help reveal autism’s biological framework and point to new avenues for therapies, she and her co-investigators say. The convergence of so many different genes on a handful of common protein interaction pathways also points to how a variety of mutations linked to autism can produce similar outcomes.
The analysis included mutations in 100 genes linked with high confidence to autism. More than 1,800 protein partners interact with the proteins encoded by those genes, Wang and her colleagues found using human embryonic kidney cells. And many of those dynamics had never been documented. “Almost 90 percent of the interaction data that we got were new,” Wang says.
P
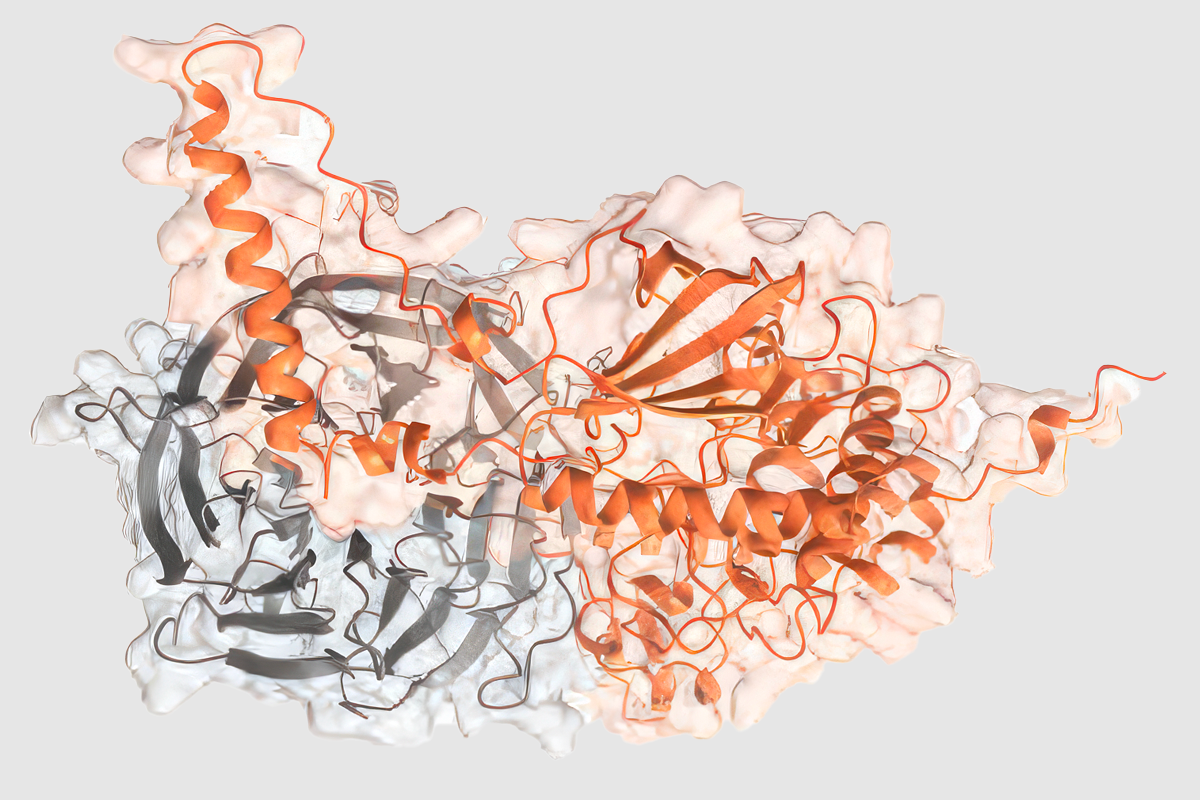
roteins encoded by autism-linked genes and their interaction partners are involved in neurogenesis, tubulin biology, transcriptional regulation and chromatin modification, the researchers found. The proteins that interact with autism-linked proteins are present in several known protein complexes.One interactor protein in particular, DCAF7, appears to bind to eight of the autism-linked proteins. And it can be found along with two of the autism-linked proteins on the cell’s division machinery, cellular studies showed.
Knocking down DCAF7 or the other two proteins in tadpoles, which have modeled the function of autism-linked genes in past work, resulted in reduced brain size. “These proteins hang out together, and they seem to have overlapping function,” Wang says.
Overall, the 100 autism-linked proteins showed an unusually high degree of connectivity with one another, says Nevan Krogan, professor of cellular and molecular pharmacology at the University of California, San Francisco, who co-led the work with Willsey. “They were more likely to fall into complexes and into pathways” than proteins linked to other conditions studied to date, he says.
The researchers then generated interaction networks for 54 specific mutations — all missense variants identified in autistic people — to see how those mutations affect protein interactions. Alphafold2, an artificial-intelligence-based protein structure prediction tool, then predicted which of the proteins in the network were likely to make direct contact with autism-linked proteins. “The direct toucher is likely to be more relevant — and ultimately a better therapeutic target as well,” Krogan says.
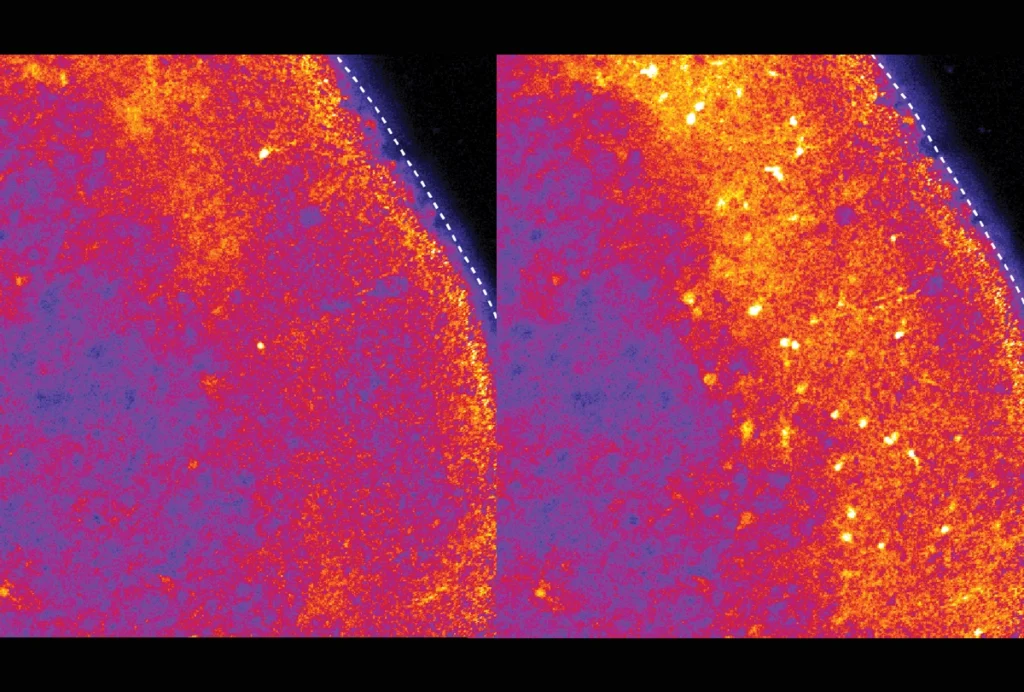
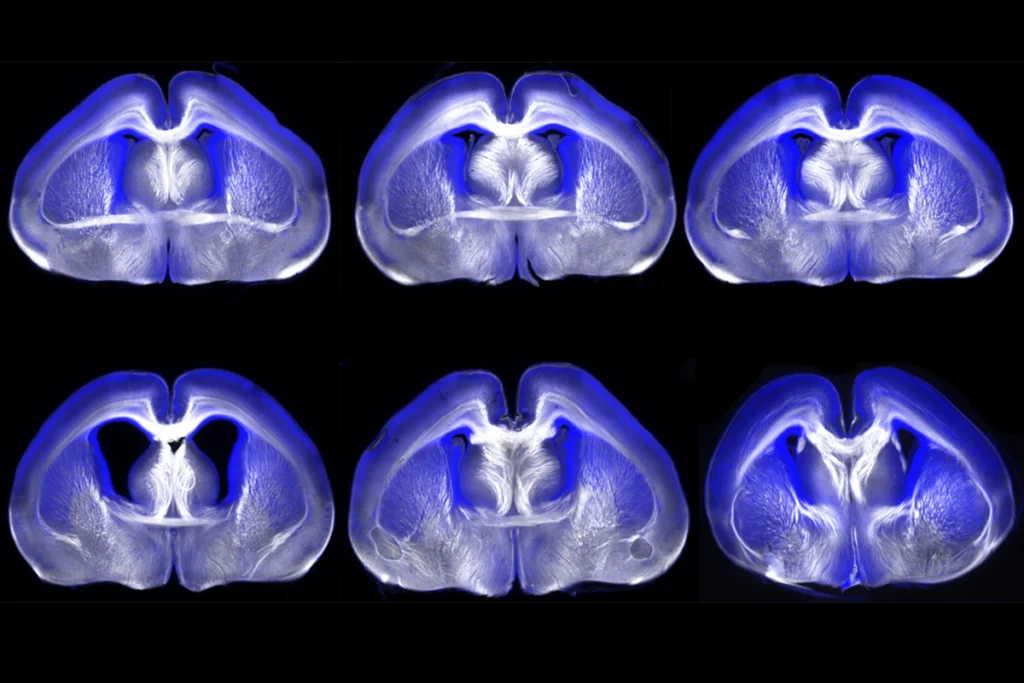
Finally, a deep-learning model homed in on FOXP1 variants as among the most probable of the 54 mutations to have a strong effect on the brain. The researchers used CRISPR to introduce a human-derived FOXP1 mutation into human induced pluripotent stem cells. Coaxing these cells to grow into human forebrain organoids revealed that this mutation alters the development of certain excitatory neurons in ways that align with differences seen in the brains of some autistic people.
E
xactly how different autism-linked genes converge functionally “is a really open question” that platforms such as this one can help address, says Kristen Brennand, professor of psychiatry and genetics at Yale University, who was not involved in this research.Overall, the approach provides a strategy for “translating those gene lists that come out of the beautiful genome studies into molecular pathway information that we can act on,” says Kasper Lage, managing director of the Novo Nordisk Foundation Center for Genomic Mechanisms of Disease at the Broad Institute, who was not involved in the study.
Some of the same interactor proteins turned up in a neuron-specific protein-interaction network that Lage and his colleagues identified last year. His team analyzed fewer genes — 13, all of which appeared on Wang and Kroger’s list — but used neurons derived from human induced pluripotent stem cells.
“Both approaches have clear advantages and disadvantages,” Lage says. Future studies should incorporate newly identified autism-linked genes and extend them to different subtypes of neurons, he adds.
“Understanding all that biological messiness is going to be important” in revealing similarities and differences between different forms of autism, he says.
Wang and her colleagues say they plan to run their analysis in neurons but are currently expanding their organoid studies to other gene mutations. They also plan to apply the same approach to other genetic conditions, such as schizophrenia. “We are just scratching the surface here,” Krogan says.