THIS ARTICLE IS MORE THAN FIVE YEARS OLD
This article is more than five years old. Autism research — and science in general — is constantly evolving, so older articles may contain information or theories that have been reevaluated since their original publication date.
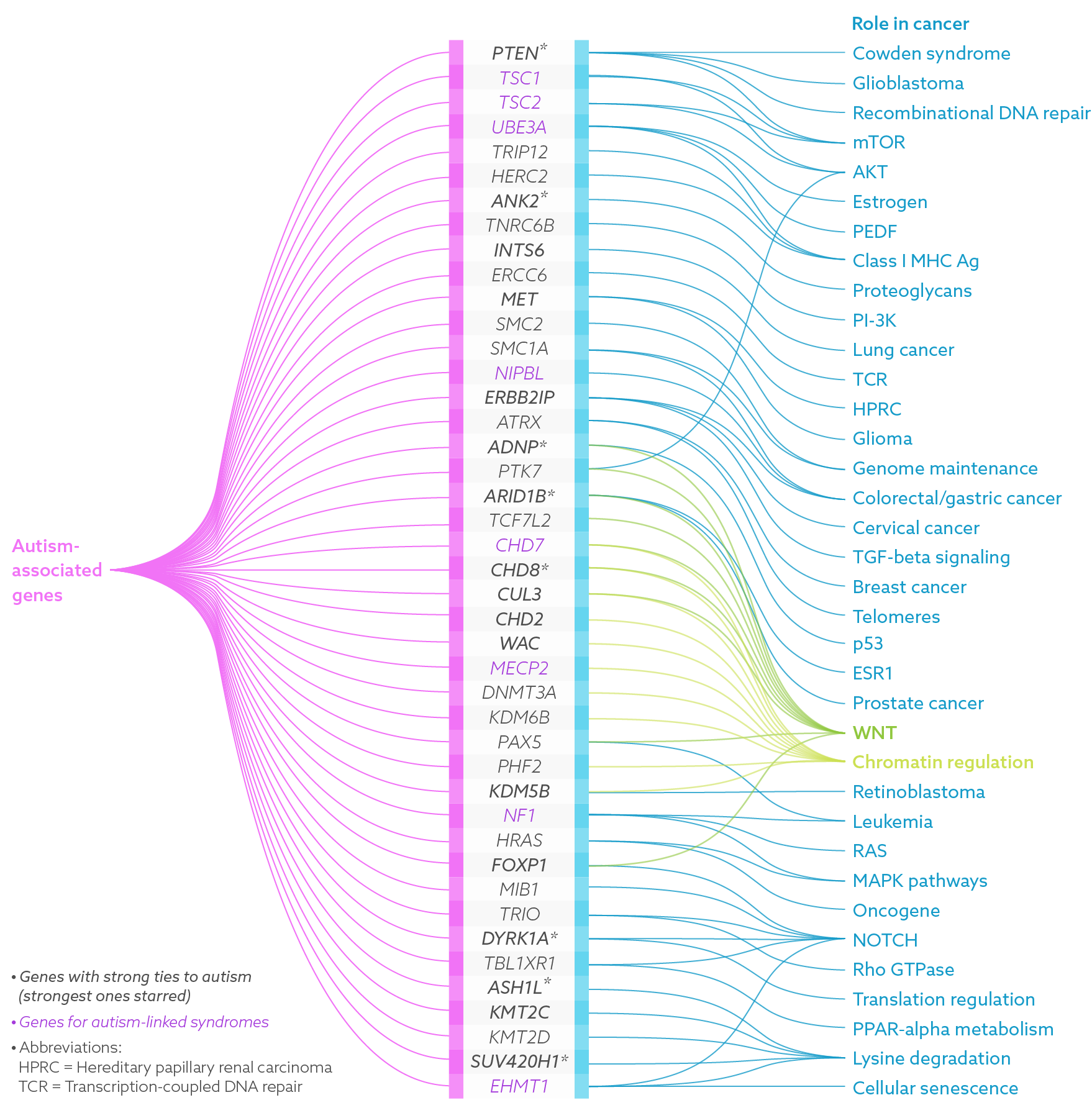
When scientists discover candidate genes for autism, they look for a compelling biological story — something that might explain how or why the gene is involved in the condition. In constructing this narrative, they’ve noted that some of the dozens of genes linked to autism are not independent actors, but cooperate with other genes.
In recent years, strong autism ties have cropped up for one group of genes in particular: those that make up a well-known signaling pathway called WNT, which also has strong links to cancer. This pathway is especially compelling because some people with autism carry mutations in various members of it, including one of its central players: beta-catenin1. What’s more, studies from the past year indicate that several of the strongest autism candidate genes, including CHD8 and PTEN, interact with this pathway.
“There might be a particular subgroup of genes associated with autism that could all be feeding into or be regulating this pathway,” says Albert Basson, reader in developmental and stem cell biology at King’s College London, who studies CHD8 and WNT. “That clearly has emerged as a relatively major theme over the last few years.”
Researchers are uncovering numerous genetic links between autism and cancer. Read more.
In April, researchers reported in Human Molecular Genetics that they had made a strain of mice that lack both copies of the beta-catenin gene. The mice show autism-like behaviors, including repetitive behaviors and difficulties with social interactions, strengthening the gene’s link to autism2.
“More and more data show that multiple genes involved in the WNT pathway can contribute to autism,” says Yingwei Mao, assistant professor of biology at Pennsylvania State University in University Park, who led the beta-catenin study. “A direction we need to consider is whether this pathway has specific functions in certain neuronal circuits.”
Separate signs:
Researchers began to suspect a biochemical link between WNT signaling and autism about 15 years ago3. This pathway governs a diverse array of functions, ranging from cell growth to the development of the brain and other organs.
In 2002, researchers made mice with a hyperactive form of beta-catenin. The animals developed enlarged brains, reminiscent of the brain overgrowth seen in some people with autism. At around the same time, epidemiological studies began to suggest that prenatal exposure to the seizure drug valproic acid — which also activates the WNT pathway — increases risk of autism in children.
A decade later, a separate team of researchers solidified the link when they uncovered hundreds of harmful mutations in people with autism but not their unaffected family members: 49 of the 126 most severe mutations occurred in a network of genes that interact with members of the WNT pathway.
This February, researchers generated mice missing two key players in the WNT pathway, the genes DVL1 and DVL34. Like mice missing beta-catenin, these mice have repetitive behaviors and impaired social interactions.
A team led by Raphael Bernier at the University of Washington in Seattle is exploring whether people with mutations in beta-catenin or in other genes in the WNT pathway similarly have features in common.
So far, the evidence indicates that mutations in any of these genes delivers a severe blow. In unpublished work presented in 2014, Bernier’s team reported that a group of 94 children with these mutations score lower on tests of intelligence and social skills than do 2,151 children with autism but no mutations in this pathway.
Rising star:
Beta-catenin rose to stardom in part because of the rise of another candidate, CHD8, that interacts with it.
In 2012, researchers flagged CHD8 as one of the top autism genes. CHD8 tightens the packing of DNA inside the nucleus. It can also interact with beta-catenin, which turns on the expression of select genes. When CHD8 interacts with beta-catenin, beta-catenin’s targets become more tightly coiled in the nucleus, preventing their expression5. So researchers hypothesize that harmful mutations in CHD8 increase beta-catenin activity.
“A lot of labs are now focused on this [pathway] because CHD8 is clearly so important in autism,” says Benjamin Cheyette, associate professor of psychiatry at the University of California, San Francisco. “That gene is really the smoking gun that’s fired up interest in WNT signaling.”
Cheyette and his colleagues have created a mouse with a mutation in one copy of CHD8. The team is exploring how mutations in CHD8 alter WNT signaling in the mice.
Others are looking at CHD8’s influence at specific points in development. For example, Anjen Chenn’s team at the University of Illinois at Chicago has made mice that lack CHD8 late in embryonic development. Loss of CHD8 increases beta-catenin activity and seems to prevent neuronal precursor cells from maturing into neurons, Chenn says.
But Cheyette says that CHD8 loss during other developmental periods may inhibit WNT signaling. “The word on the street is that whether CHD8 activates or inhibits WNT signaling targets is context dependent — sometimes it inhibits and sometimes it activates,” he says. “In other words, it’s complicated.”
Puzzle pieces:
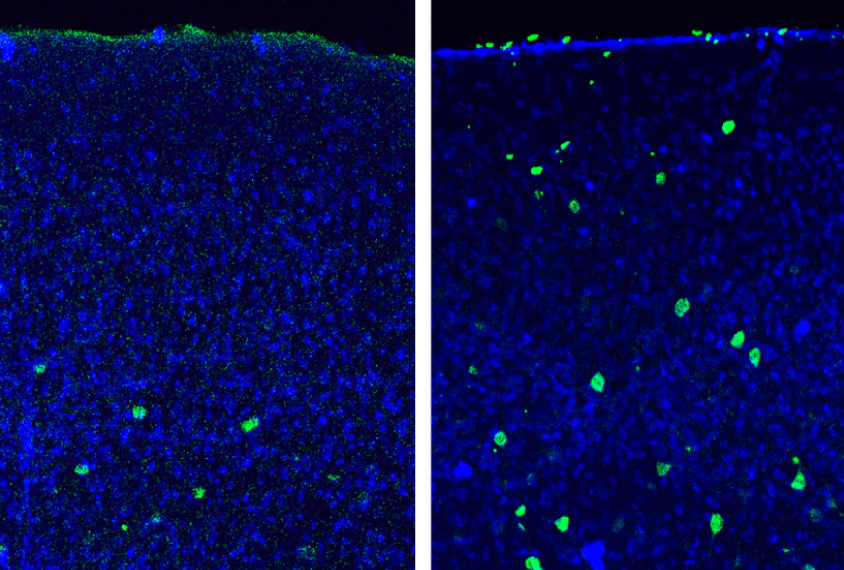
Dissecting the role of WNT signaling in particular cell types at different stages of brain development may help clarify this complexity. In the April beta-catenin study, researchers created mice that lack beta-catenin shortly after birth and in only one type of neuron: parvalbumin interneurons. These neurons dampen brain activity and are thought to be dysfunctional in the brains of people with autism.
The mice have several autism-like behaviors. They show no preference for an unfamiliar mouse over a familiar one, a behavior reminiscent of the social difficulties in people with autism. Compared with controls, they also spend more time grooming themselves, are more anxious, and show problems with long-term memory.
The findings suggest that loss of beta-catenin function only in parvalbumin interneurons and at that stage of development is enough to trigger autism-like behaviors.
“We think these inhibitory neurons might be contributing to the social interaction deficit or the repetitive behavior,” says Mao.
The mice also have an increased number of parvalbumin interneurons, suggesting that too much inhibition in the brain contributes to the autism-like behaviors, but it’s unclear how beta-catenin and WNT signaling are involved.
“It would be interesting to see, at the level of circuit activity, and specifically synapses, how the increased number of parvalbumin cells leads to these behavioral changes,” says Damon Page, associate professor of neuroscience at the Scripps Research Institute in Jupiter, Florida, who was not involved in the work.
Page’s team has found a link between beta-catenin and PTEN, another top autism candidate that also has ties to cancer. In 2015, his group reported that mice lacking a copy of PTEN develop enlarged brains through a mechanism that requires beta-catenin6. The team is now trying to figure out how loss of PTEN and beta-catenin, individually and together, contribute to brain development and autism-like behaviors.
By assembling these disparate pieces of data, researchers hope to get a full picture of the role of WNT signaling in autism.
“We have a puzzle that’s very complicated, with a million jigsaw pieces that need to be fit together,” says Cheyette. “If you’ve ever done a jigsaw puzzle, what do you do? You start out working on just a little corner, you try and fit a few of the pieces together, maybe you work on the edges and you fill in the center bit by bit.”
By joining the discussion, you agree to our privacy policy.